Using Comparative Genomics to Identify New Conserved, Sexual Reproduction Genes
With Dr. Oliver Billker and colleagues, we extended our comparative studies of sexual reproduction in Chlamydomonas and Plasmodium. We used RNA-Seq methods to determine transcript abundance in Chlamydomonas cells in eight different physiological states. Hierarchical cluster analysis grouped all known sexual reproduction genes in closely related clusters. Using this approach we identified our second conserved gene, GEX1, which functions at similar steps in sexual reproduction in Chlamydomonas, Plasmodium, and higher plants. As with HAP2, disruption of GEX1 blocks proliferation of meiotic progeny in both protists. Because Plasmodium meiosis occurs in the gut of the mosquito, Plasmodium GEX1 is essential for mosquito transmission of malaria. (Ning et al., 2013).
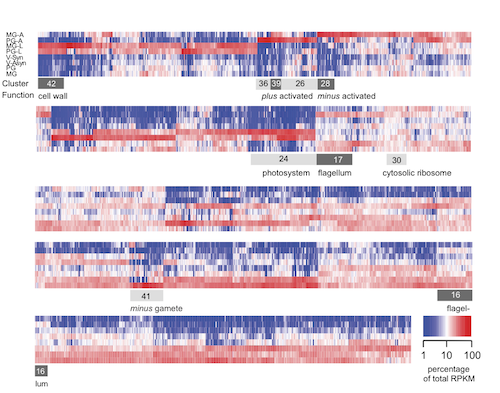
Cluster analysis of gene expression patterns in Chlamydomonas gametes and vegetative cells
identifies sexual reproduction genes
Remarkably, we discovered that the GEX1 genes are members of a large, previously unrecognized family, whose first-characterized member, KAR5, is essential for nuclear fusion during yeast sexual reproduction. We show that the GEX1/KAR5 family also includes a recently reported nuclear envelope protein essential for nuclear fusion and for early embryo formation in zebrafish.