Movement of RNA between animal cells and across generations
Generation after generation, we begin life as a single cell from which arise many different cell types that act together seamlessly and respond to our environment. How does one cell coordinate its function with another cell? Does what our body cells experience during one generation affect subsequent generations? To answer these long-standing questions, we use the simple worm C. elegans to reveal mechanisms that convey regulatory information from one cell to another and from one generation to the next.
RNA as a carrier of regulatory information form somatic cells to germ cells: Double-stranded RNA (dsRNA) can silence genes of matching sequence in many organisms, including humans. 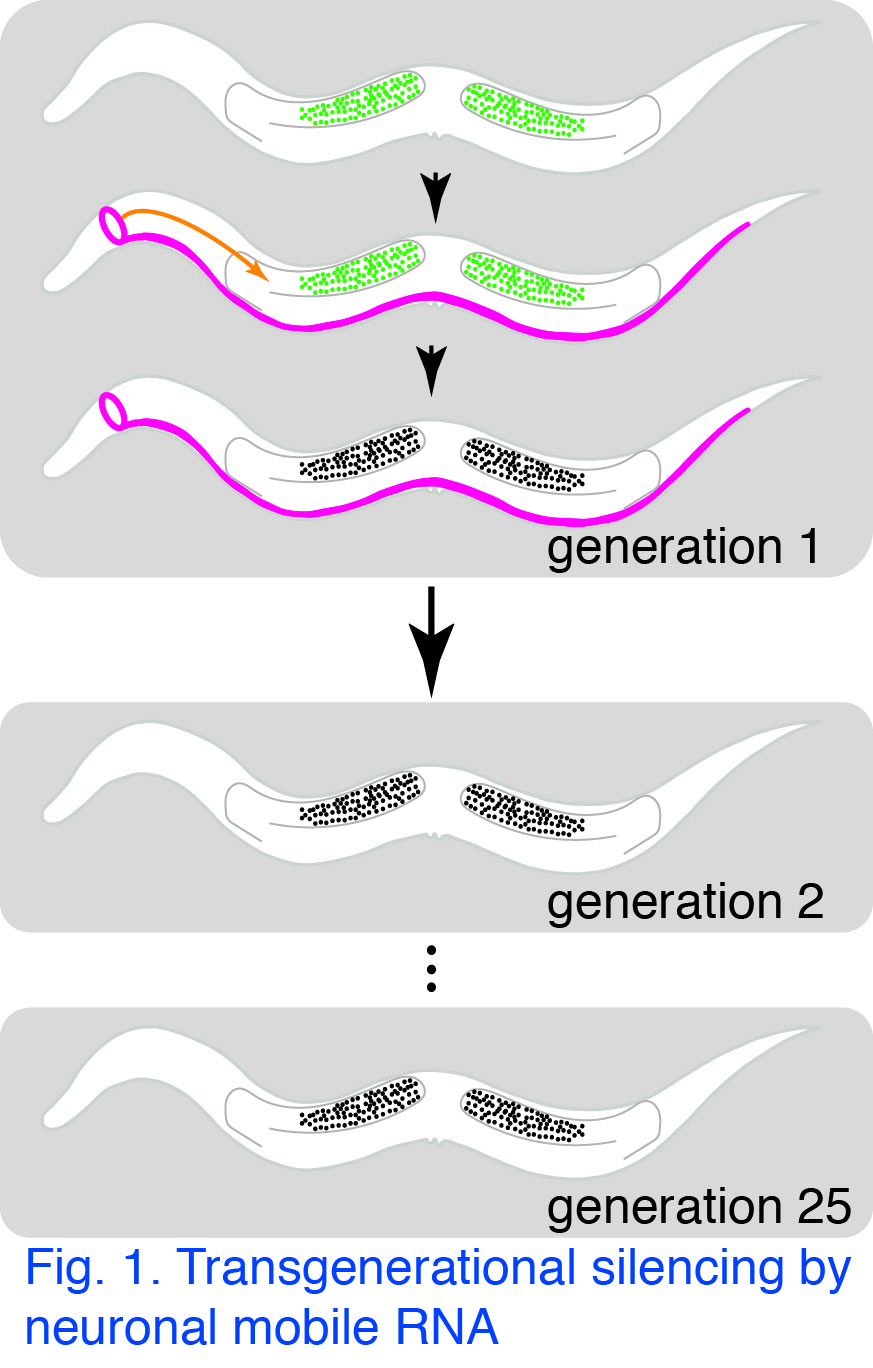 Using the simple worm C. elegans, we found that dsRNA made in neurons can enter the germline and cause transgenerational gene silencing (Fig. 1). Neurons (magenta) can export (orange arrow) forms of double-stranded RNA called mobile RNA that match a gene (green) in germ cells. Import of RNA into germ cells results in silencing of the gene (black) within germ cells. This silencing can persist for more than 25 generations in the absence of the ancestral source of mobile RNA. Our goal is to understand how this remarkable intercellular transport of mobile RNA and the resultant transgenerational silencing are accomplished.
Using the simple worm C. elegans, we found that dsRNA made in neurons can enter the germline and cause transgenerational gene silencing (Fig. 1). Neurons (magenta) can export (orange arrow) forms of double-stranded RNA called mobile RNA that match a gene (green) in germ cells. Import of RNA into germ cells results in silencing of the gene (black) within germ cells. This silencing can persist for more than 25 generations in the absence of the ancestral source of mobile RNA. Our goal is to understand how this remarkable intercellular transport of mobile RNA and the resultant transgenerational silencing are accomplished.
Intergenerational and Transgenerational Inheritance: A broad framework to understand intergenerational inheritance (from one generation to the next) and transgenerational inheritance (across many generations) has emerged from studies in many model systems. Models for both forms of inheritance hinge on the ability of small RNAs to direct chromatin and DNA modifications by base-pairing with nascent RNA of matching sequence. Current models for intergenerational silencing in C. elegans propose that entry of dsRNA into germ cells results in the production of secondary small RNAs through the action of RNA-dependent RNA polymerases and it is these secondary RNAs and/or RNA-dependent chromatin modifications that are inherited from one generation to the next. Current models for transgenerational inheritance in C. elegans propose that stable chromatin, possibly associated with the production of small RNAs that reinforce these modifications in each generation, are inherited across many generations. Most aspects of this framework, however, remain to be tested.
Movement of RNA between somatic cells: Nearly organism-wide spread of gene silencing is observed when dsRNA is injected into one tissue in C. elegans, suggesting that mobile RNAs derived from these dsRNAs spread to other somatic tissues. Indeed, multiple cases of inter-tissue movement of mobile RNAs derived from expressed dsRNA have been observed: from neurons to gut cells, from muscle cells to gut cells, and from gut cells to muscle cells. However, somatic silencing by mobile RNAs appears to not trigger transgenerational silencing. For example, although transgenerational silencing by neuronal mobile RNAs that enter germ cells is easily detected, such inherited silencing by mobile RNAs that enter somatic cells is not detectable. These observations raise many questions. For example, are mobile RNAs that enter germ cells the same as those that enter somatic cells? Are mobile RNAs derived from dsRNA injected into a tissue the same as those derived from dsRNA expressed within a tissue? Nevertheless, it is clear that mechanisms that export and import mobile RNAs are present in many somatic cell types.
Mechanisms of RNA transport between cells: The detection of circulating RNAs in humans and the potential use of such RNAs as diagnostics has ignited interest in RNA transport between animal cells. Multiple mechanisms of specific secretion have been proposed to account for circulating RNAs with differing degrees of experimental support. RNAs protected within vesicles called exosomes and RNAs stabilized by RNA-binding proteins have emerged as the most popular mechanisms that also account for the stability of these RNAs in the extracellular environment. The use of genetic mutants and animal models will be essential to gain more confidence in each of the proposed RNA transport mechanisms and to connect the presence of circulating RNAs with specific biological function. Therefore, we are using the genetically tractable animal model C. elegans to analyze RNA transport mechanisms - specifically, those that transport mobile RNAs between cells.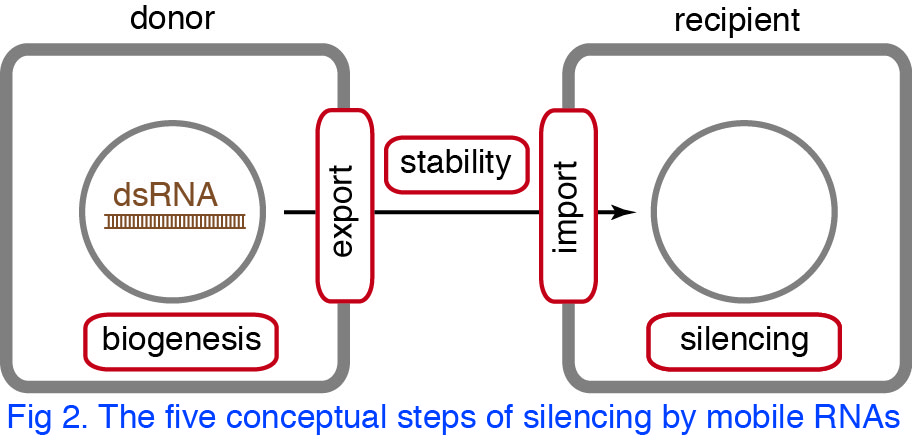
Silencing by mobile RNAs can be conceptually divided into five steps (Fig. 2). Mobile RNAs need to be made from transcribed dsRNA (biogenesis), secreted from donor cells (export), protected in extracellular space (stability), and imported into recipient cells (import) to silence the target gene (silencing). Genetic screens have the potential to uncover the molecular machinery that is required for each of these steps.
The analysis of genes identified through previous genetic screens in C. elegans and their mammalian homologs has provided insight into RNA import mechanisms. We now know that the import of dsRNA into C. elegans cells requires a conserved dsRNA-selective transporter protein and is stimulated by signaling through a conserved tyrosine kinase. Analysis of additional conserved genes from a screen in D. melanogaster and of other C. elegans-specific genes has lead to a basic understanding of the import of dsRNA into cells. Current models for the import of dsRNA propose an endocytic uptake of dsRNA followed by release into the cytosol through either the dsRNA-selective importer protein in C. elegans and in mammals or through as yet unclear mechanisms in Drosophila.
We have some clues about the biogenesis of mobile RNAs in C. elegans. The requirement for the dsRNA-selective importer in C. elegans suggests that mobile RNAs are likely forms of dsRNA. The same conclusion was also reached using genetic mosaic analyses of animals with RNA silencing factors expressed in donor or recipient cells. Despite these two independent lines of evidence, several questions remain. For example, are mobile RNAs chemically modified forms of dsRNA? What protein factors, if any, perform such modifications?
Finally, how mobile RNAs are exported, what factors, if any, are required for their stability in the extracellular space, and if silencing by imported mobile RNAs differs from silencing by dsRNA made within cells, remain unknown.
To identify additional genes required for silencing by mobile RNAs, we generated a sensor strain that expresses dsRNA against green fluorescent protein (gfp) under a neuronal promoter in an enhanced RNA silencing background (eri-1(-)), which results in potent silencing of gfp in non-neuronal cells. Using this sensor strain, we have identified many mutants called silencing by eri-1 loss or by neuronal dsRNA defective (send) mutants. We expect that the analysis of the corresponding send genes and genes identified through additional genetic screens will shed more light on the mechanisms that control silencing by mobile RNAs.
Perspective: The movement of mobile RNAs between cells transforms our understanding of how cells of an animal regulate each other and possibly how animals evolve. Typically, a cell interprets extracellular signals. For example, when a skeletal muscle cell binds acetylcholine, it increases contraction, but when a salivary gland cell binds acetylcholine, it increases secretion. In contrast, if a mobile RNA enters a cell, the gene with matching sequence is shut off without any interpretation by the cell. Consequently, mobile RNAs that move from body cells to germ cells in a certain environment can convey gene-specific information from one generation to the next. Inheritance of such information can accelerate the evolution of multicellular organisms.
Back to research
Last updated: Dec 2015