Electrophoresis
Page
history edited by davisvas@gmail.com
…revised
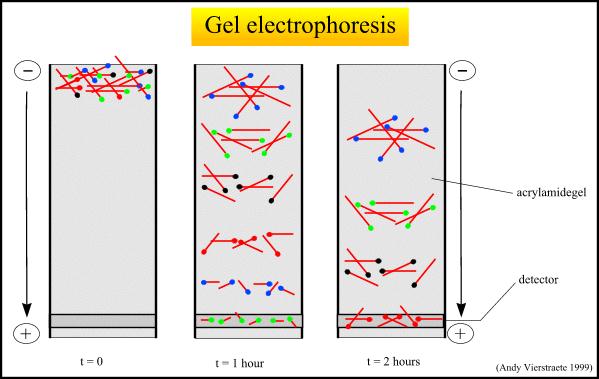
Electrophoresis
can be used to sort DNA segments by size. DNA segments are placed
at one end of a gel. DNA is negatively charged (with a charge of two
electrons per base pair). Then an electric field is applied through
connecting anodes and cathodes at the ends of the gel. This causes the
negative DNA segments to move towards the positive end of the gel. After
a time, the field is turned off and the DNA visualized. The smaller DNA segments have moved farther
from the starting end. Why do the smaller DNA segments move farther?
1. Each base pair of a DNA molecule has a
negative charge of -2 elementary charges. Defining whatever variables you
find appropriate, come up with an expression for:
a) the total
charge of a DNA segment of N
base pairs
b) the total mass
of a DNA segment of N
base pairs
c) the total
length of a DNA segment of N
base pairs
2. To understand how electrophoresis
works, let's first start with the simplest possible setup: let's ignore
the gel and assume that the experiment is taking place in a vacuum, so the only
force acting on the DNA segments is the electrostatic force due to a constant
electric field.
a) Using whatever representational tools
are useful (free-body diagrams, equations, etc.), model how this electrostatic
force acts on DNA segments of different sizes. (Does this force have a
different effect on different-sized segments of DNA?)
b) Does this explain how electrophoresis
separates DNA segments by size?
3. Obviously, a gel is not a vacuum!
So now let's include the effects of the gel. Let's consider drag forces
that are proportional to the cross-sectional area (as we did during the first
semester). Once again, model how the forces (electrostatic and drag) act
on DNA segments of different sizes. Again, do these forces have different
effects on different-sized segments of DNA? Your findings may depend on
your assumptions about geometry, so try it for both
a) assuming that
the DNA segment is basically shaped like a long straight rope (as illustrated
in the picture below)
b) assuming that
the DNA segment curls up into (approximately) a sphere.
(Hint: Think about the volume of a
sphere made out of a given number of base pairs where each base pair has the same
mass, and then think about the cross-sectional area of that sphere.)
(Feel
free to try out any other geometries too!)
4. Given
that we know electrophoresis is used to separate DNA segments by size,
with smaller
segments moving farther in a given amount of time, have we
come up with a model that explains this result? If not, what assumptions
that we have made need to be reconsidered? Is there another model that
can account for this result?
5. When
DNA segments are very long the separation becomes poor or there is no
separation at all. What could explain
this finding? (You need to think outside
the box for this one…or look this up. It’s
important to realize that the mechanics of moving macromolecules through a gel
is more complex than this fundamental analysis would predict.)