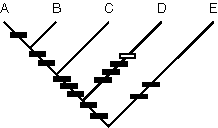
1 10 20
|........|.........|....
Aster GTCATACCTGTGACATAGGTAATT
Barnadesia ATCATACCTGTGACATAGGTAATT
Cornus ATCTTCCCTGTGACATAGGTAATT
Dioscorea ATCTTACCGGTTACTTACGCCATT
Ephedra ATCTTCCCGGTTACTCTGGTAAGG