
ZhongchiLiu Lab Research Interests
 We are interested in how cells in a multicellular
organism assume their developmental fates and form distinct
patterns. We have chosen to address this fundamental question
in the context of Arabidopsis flower development by focusing
on the regulatory mechanism of the floral homeotic gene AGAMOUS
(AG) . AG is a key regulatory gene for flower
development with at least three functions: the specification
of carpel and stamen identity, the repression of sepal and
petal identity, and the control of floral meristem determinacy.
Previous studies established that the activity of AG
is mainly regulated at the transcription level. Negative
regulation of AG transcription is crucial in ensuring stage-
and tissue-specific AG mRNA expression and hence proper
floral organ identity and floral organ pattern. However, little
is known about the mechanism underlying AG transcriptional repression.
We are interested in how cells in a multicellular
organism assume their developmental fates and form distinct
patterns. We have chosen to address this fundamental question
in the context of Arabidopsis flower development by focusing
on the regulatory mechanism of the floral homeotic gene AGAMOUS
(AG) . AG is a key regulatory gene for flower
development with at least three functions: the specification
of carpel and stamen identity, the repression of sepal and
petal identity, and the control of floral meristem determinacy.
Previous studies established that the activity of AG
is mainly regulated at the transcription level. Negative
regulation of AG transcription is crucial in ensuring stage-
and tissue-specific AG mRNA expression and hence proper
floral organ identity and floral organ pattern. However, little
is known about the mechanism underlying AG transcriptional repression.

 We have isolated and characterized three key negative
regulators of AG. They are LEUNIG (LUG)
, SEUSS (SEU) and LARSON
(LSN). We showed that the homeotic transformation
in the floral organ identity of lug
and seu mutants was mediated by precocious and ectopic
AG expression (Liu and Meyerowitz, 1995; Franks
et al., 2002). Further, using a map-based approach,
we have isolated LUG and SEU.
LUG encodes a putative transcriptional
co-repressor with sequence similarity to a class of
transcription co-repressors including Tup1 from
yeast, Groucho from Drosophila, and TLE from mammals
(Conner and Liu, 2000). SEU encodes a novel
protein with at least two glutamine-rich domains
and a conserved domain that shares sequence identity with the
dimerization domain of the LIM-domain-binding transcription
co-regulators in animals (Franks et al., 2002). We
are actively testing the hypothesis that LUG
and SEU may form a co-repressor complex, that is recruited
to the AG cis-regulatory element by other DNA-binding
transcription factors that bind to AG cis-regulatory
elements.
We have isolated and characterized three key negative
regulators of AG. They are LEUNIG (LUG)
, SEUSS (SEU) and LARSON
(LSN). We showed that the homeotic transformation
in the floral organ identity of lug
and seu mutants was mediated by precocious and ectopic
AG expression (Liu and Meyerowitz, 1995; Franks
et al., 2002). Further, using a map-based approach,
we have isolated LUG and SEU.
LUG encodes a putative transcriptional
co-repressor with sequence similarity to a class of
transcription co-repressors including Tup1 from
yeast, Groucho from Drosophila, and TLE from mammals
(Conner and Liu, 2000). SEU encodes a novel
protein with at least two glutamine-rich domains
and a conserved domain that shares sequence identity with the
dimerization domain of the LIM-domain-binding transcription
co-regulators in animals (Franks et al., 2002). We
are actively testing the hypothesis that LUG
and SEU may form a co-repressor complex, that is recruited
to the AG cis-regulatory element by other DNA-binding
transcription factors that bind to AG cis-regulatory
elements.
LSN appears to encode an excellent candidate DNA-binding
partner of the LUG/SEU co-repressor.
lsn mutations enhance the defects of lug
, and LSN encodes a putative transcription factor
with a DNA-binding motif (Xiaozhong Bao and Zhongchi Liu, unpublished
result). Further molecular analyses of how LSN, LUG
and SEU interact will provide important insights into
the repression mechanism of floral homeotic geneexpression.
 in situ hybridization showing AG expression
in a wild-type inflorescence.
in situ hybridization showing AG expression
in a wild-type inflorescence.
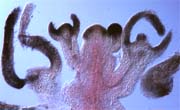 AG expression in seu-1 lug-1 double mutant
inflorescence.
AG expression in seu-1 lug-1 double mutant
inflorescence.
A second emphasis in the lab was
the genetic and molecular characterization
of tso1 and tso2 mutants. We are
interested in understanding how inflorescence and floral
meristems are organized and what genes regulate the cell division
activity and cell division orientation in inflorescence
and floral meristems. Mutations in TSO1 cause
defects in cell division in Arabidopsis floral
meristems and cause inflorescence meristem fasciation. We
have isolated the TSO1 gene using the map-based
approach and showed that TSO1 encodes two putative
DNA-binding cysteine-rich repeats and is localized
to the nucleus (Song et al., 2000). Thus,
TSO1 likely encodes a transcriptional factor
that regulates the expression of genes involved in cell division
and meristem organization. Our lab is further characterizing
the TSO1 protein, its localization, and the regulation
of its expression. We have a postdoctoral position open for
this project. If interested, please contact Dr. Zhongchi Liu
via email ( ZL17@umail.umd.edu
).
How are the genes named?
LEUNIG was named after the famous Australian catoonist
Michael Leunig because
of the resemblance of the horn-like protrutions at the
tip of leunig mutant gynoecium to the horn-like protruions
in Michael Leunig's cartoon characters.
Dr. David Smyth was responsible for naming
the gene.
SEUSS and LARSON were both initially identified
by mutations that enhance leunig, we decided
to name them following the tradition of naming after cartoonist.
Bob Franks (a former postdoctoral fellow in the lab) decided
to name the two genes after the American cartoonists
Dr. SEUSS and
Gary Larson.
TSO1 and TSO2 genes are named so because of the ugly floral
morphology in tso1 and tso2 mutants.
TSO means ugly in Chinese.
Current Lab members
 Xiaozhong Alex Bao, graduate
student, xbao@wam.umd.edu
Xiaozhong Alex Bao, graduate
student, xbao@wam.umd.edu
 Jayashree Sitaraman, graduate
student, jais@wam.umd.edu
Jayashree Sitaraman, graduate
student, jais@wam.umd.edu
 V. V. Srihdar, postdoctoral
fellow, vvsridhar65@yahoo.com
V. V. Srihdar, postdoctoral
fellow, vvsridhar65@yahoo.com
 Anandkumar Surendrarao, graduate
student, sak@wam.umd.edu
Anandkumar Surendrarao, graduate
student, sak@wam.umd.edu
 Chunxing Wang, graduate student,
cxwang@wam.umd.edu
Chunxing Wang, graduate student,
cxwang@wam.umd.edu
Some Former Lab members
 Bob Franks, postdoctoral fellow,
rgfranks@uclink.berkeley.edu
Bob Franks, postdoctoral fellow,
rgfranks@uclink.berkeley.edu
Joann Conner,
postdoctoral fellow
Jaiyoung Song, postdocteral fellow
Terri Leung, technician,
tsleung00@yahoo.com
Some lab pictures

Lab picture, 1999 fall,
in front of the H.J. Patterson Hall. From left: Terri
Leung, Sarah Brooks, Bob Franks, Seth Glatstein, Jai-Young
Song, Black Wang, and Zhongchi Liu

Lab
picture, 2000 spring, at Dumbartom
Oaks , Washington D.C. From left,
Anandkumar Surendrarao, Zhongchi Liu, Black Wang, Alex Bao, and Bob
Franks.

Lab picture, 2000 summer, at
the Hirshhorn Sculpture
Garden, Smithsonian, Washington D.C. From front to back, Anandkumar
Surendrarao, Black Wang, and Bob Franks.
Local Group meetings
GEMS (
Genetics with Eukaryotic Model Systems): a monthly group gethering
among local researchers who study eukaryotic model systems such as
Arabidopsis, Drosophila and C. elegans.
Atrium (Arabidopsis thaliana Research Initiative
at University of Maryland): a monthly group meeting among
local researchers who study Arabidopsis. An anual mini-symposium
is held in the spring of each year.
Course Development
BSCI 411 Plant Genetics and Molecular Biology
BSCI 348G Laboratory in Molecular Genetics
MICB688G/PBIO699G
Genetic Approaches to Cell and Developmental Biology
Recent Publications
Franks, R. Wang, C., Levin, J. Z., and Liu, Z. (2002) SEUSS,
a member of a novel family of
plant regulatory proteins, represses floral homeotic
gene expression with LEUNIG . Development 129: 253-263.
( click here to download
the pdf file) (with Cover ).
Franks, R.G. and Liu, Z. (2001) Floral homeotic gene regulation.
Horticultural Reviews vol. 27, 41-77.
Song J, Leung T, Ehler LK, Wang C, Liu Z. (2000) Regulation of meristem
organization and cell division
by TSO1 , an Arabidopsis gene with cysteine-rich
repeats. DEVELOPMENT 127:2207-2217. (
pdf/fulltext )
Liu, Z., Franks, R.G. and Klink, V. P. (2000) Regulation of
marginal tissue formation by LEUNIG and AINTEGUMENTA
. PLANT CELL 12, 1893-1902. (with
Cover ).
Conner, J. and Liu Z. (2000) LEUNIG , a putative
transcriptional co-repressor
that regulates AGAMOUS expression during flower
development. PNAS 97, 12902-12907.
(click here to download the
pdf file) .
Scovel, G., Altshuler, T., Liu, Z., and Vainstein, A. (2000)
The EVERGREEN gene is essential for flower
initiation in carnation. J. of Heredity 91,
487-491.
Liu, Z., Running, M. R., and Meyerowitz, E. M. (1997) TSO1
function in cell division during Arabidopsis flower development,
DEVELOPMENT 124: 665-672. (
pdf/fulltext )
Liu, Z., and Meyerowitz E. M. (1995). LEUNIG regulates
AGAMOUS expression in Arabidopsis flowers.
DEVELOPMENT 121: 975-991.
 We are interested in how cells in a multicellular
organism assume their developmental fates and form distinct
patterns. We have chosen to address this fundamental question
in the context of Arabidopsis flower development by focusing
on the regulatory mechanism of the floral homeotic gene AGAMOUS
(AG) . AG is a key regulatory gene for flower
development with at least three functions: the specification
of carpel and stamen identity, the repression of sepal and
petal identity, and the control of floral meristem determinacy.
Previous studies established that the activity of AG
is mainly regulated at the transcription level. Negative
regulation of AG transcription is crucial in ensuring stage-
and tissue-specific AG mRNA expression and hence proper
floral organ identity and floral organ pattern. However, little
is known about the mechanism underlying AG transcriptional repression.
We are interested in how cells in a multicellular
organism assume their developmental fates and form distinct
patterns. We have chosen to address this fundamental question
in the context of Arabidopsis flower development by focusing
on the regulatory mechanism of the floral homeotic gene AGAMOUS
(AG) . AG is a key regulatory gene for flower
development with at least three functions: the specification
of carpel and stamen identity, the repression of sepal and
petal identity, and the control of floral meristem determinacy.
Previous studies established that the activity of AG
is mainly regulated at the transcription level. Negative
regulation of AG transcription is crucial in ensuring stage-
and tissue-specific AG mRNA expression and hence proper
floral organ identity and floral organ pattern. However, little
is known about the mechanism underlying AG transcriptional repression.


 We have isolated and characterized three key negative
regulators of AG. They are LEUNIG (LUG)
, SEUSS (SEU) and LARSON
(LSN). We showed that the homeotic transformation
in the floral organ identity of lug
and seu mutants was mediated by precocious and ectopic
AG expression (Liu and Meyerowitz, 1995; Franks
et al., 2002). Further, using a map-based approach,
we have isolated LUG and SEU.
LUG encodes a putative transcriptional
co-repressor with sequence similarity to a class of
transcription co-repressors including Tup1 from
yeast, Groucho from Drosophila, and TLE from mammals
(Conner and Liu, 2000). SEU encodes a novel
protein with at least two glutamine-rich domains
and a conserved domain that shares sequence identity with the
dimerization domain of the LIM-domain-binding transcription
co-regulators in animals (Franks et al., 2002). We
are actively testing the hypothesis that LUG
and SEU may form a co-repressor complex, that is recruited
to the AG cis-regulatory element by other DNA-binding
transcription factors that bind to AG cis-regulatory
elements.
We have isolated and characterized three key negative
regulators of AG. They are LEUNIG (LUG)
, SEUSS (SEU) and LARSON
(LSN). We showed that the homeotic transformation
in the floral organ identity of lug
and seu mutants was mediated by precocious and ectopic
AG expression (Liu and Meyerowitz, 1995; Franks
et al., 2002). Further, using a map-based approach,
we have isolated LUG and SEU.
LUG encodes a putative transcriptional
co-repressor with sequence similarity to a class of
transcription co-repressors including Tup1 from
yeast, Groucho from Drosophila, and TLE from mammals
(Conner and Liu, 2000). SEU encodes a novel
protein with at least two glutamine-rich domains
and a conserved domain that shares sequence identity with the
dimerization domain of the LIM-domain-binding transcription
co-regulators in animals (Franks et al., 2002). We
are actively testing the hypothesis that LUG
and SEU may form a co-repressor complex, that is recruited
to the AG cis-regulatory element by other DNA-binding
transcription factors that bind to AG cis-regulatory
elements. in situ hybridization showing AG expression
in a wild-type inflorescence.
in situ hybridization showing AG expression
in a wild-type inflorescence. AG expression in seu-1 lug-1 double mutant
inflorescence.
AG expression in seu-1 lug-1 double mutant
inflorescence.  Xiaozhong Alex Bao, graduate
student, xbao@wam.umd.edu
Xiaozhong Alex Bao, graduate
student, xbao@wam.umd.edu Jayashree Sitaraman, graduate
student, jais@wam.umd.edu
Jayashree Sitaraman, graduate
student, jais@wam.umd.edu V. V. Srihdar, postdoctoral
fellow, vvsridhar65@yahoo.com
V. V. Srihdar, postdoctoral
fellow, vvsridhar65@yahoo.com Anandkumar Surendrarao, graduate
student, sak@wam.umd.edu
Anandkumar Surendrarao, graduate
student, sak@wam.umd.edu Chunxing Wang, graduate student,
cxwang@wam.umd.edu
Chunxing Wang, graduate student,
cxwang@wam.umd.edu
 Bob Franks, postdoctoral fellow,
rgfranks@uclink.berkeley.edu
Bob Franks, postdoctoral fellow,
rgfranks@uclink.berkeley.edu

