Methods
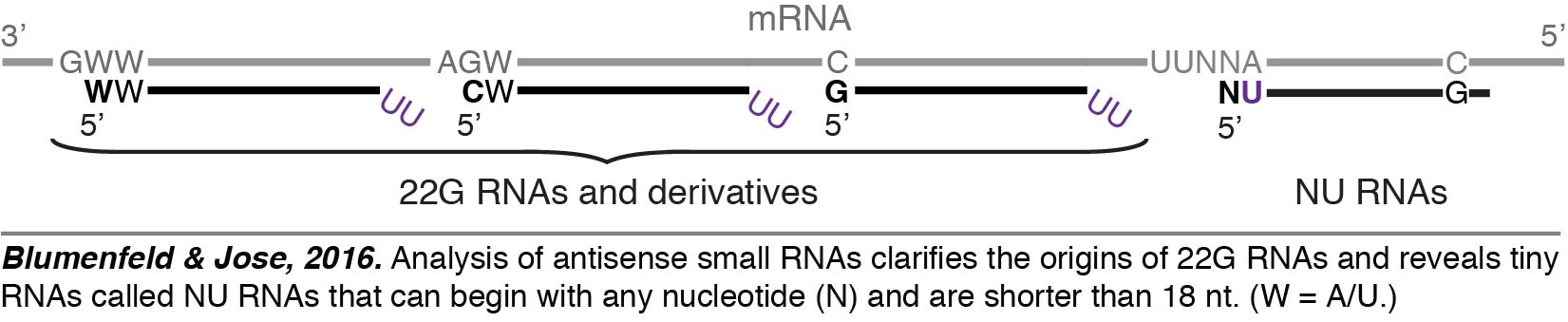
Analysis of small RNAs. Small RNAs are major regulators of gene expression in C. elegans, and there are millions of them. They are typically identified using RNA-seq, but the effective analysis of the resulting data is a challenge. Therefore, we created a set of programs called PACER (Programs for Analysis of C. elegans small RNAs) to effectively analyze small RNAs detected through RNA-seq (Blumenfeld & Jose, 2016). Using PACER, we gained insight into the origins of the diverse set of antisense small RNAs called 22G RNAs that are used for sequence-specific gene regulation in C. elegans and discovered a new class of tiny RNAs that are shorter than 18 nucleotides (18-nt) called NU RNAs (pronounced “new RNAs”). The minimum length of a small RNA that is required for sequence-specific regulation depends on the sequence space that needs to be searched. Thus, we propose that RNAs shorter than 18-nt could be used for sequence-specific regulation – particularly in organisms with small genomes – and represent an underexplored world of functional tiny RNAs. This speculation is supported by our detection of NU RNAs despite most RNA-seq experiments selecting against such RNAs.
Sensitive northern blotting. When analyzing RNA-seq data, we discovered that some of the sequences present in a dataset could be generated during the preparatory steps required for RNA-seq, 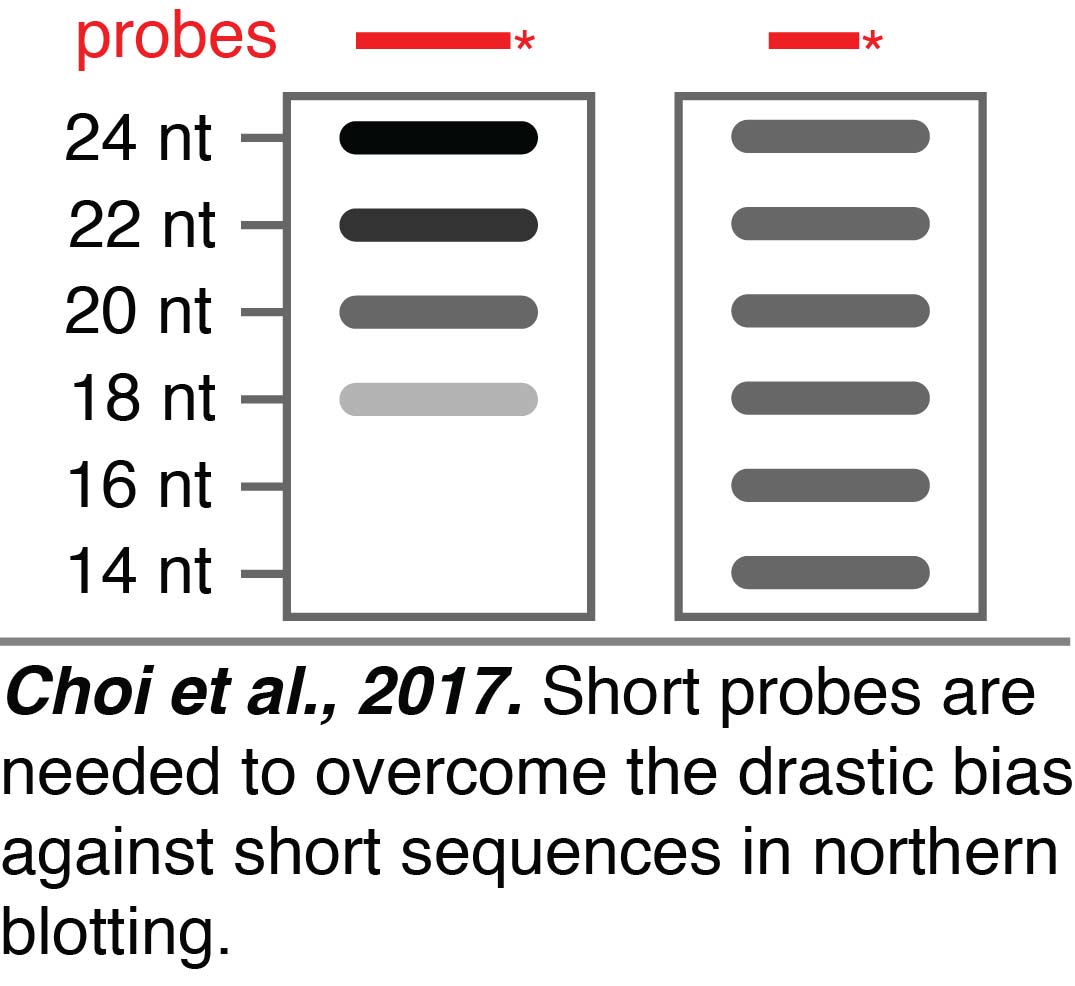 highlighting the need for additional approaches to study small RNAs. Small RNAs can be independently examined using northern blotting. We found that northern blotting of small RNAs can have a drastic bias against short sequences (Choi et al., 2017). We greatly reduced the bias from ~360 fold against 18 nt RNAs compared to 24 nt RNAs when using a 24 nt probe to a maximum of ~4 fold across the entire range of 24 nt to 14 nt RNAs by using 15 nt probes and low hybridization temperatures. This improved northern blotting will be useful for the independent evaluation of any tiny RNAs that we discover when we perform RNA-seq without selecting against RNAs that are less than 18-nt.
highlighting the need for additional approaches to study small RNAs. Small RNAs can be independently examined using northern blotting. We found that northern blotting of small RNAs can have a drastic bias against short sequences (Choi et al., 2017). We greatly reduced the bias from ~360 fold against 18 nt RNAs compared to 24 nt RNAs when using a 24 nt probe to a maximum of ~4 fold across the entire range of 24 nt to 14 nt RNAs by using 15 nt probes and low hybridization temperatures. This improved northern blotting will be useful for the independent evaluation of any tiny RNAs that we discover when we perform RNA-seq without selecting against RNAs that are less than 18-nt.
Epigenetic sensor. We developed a drug-sensitive epigenetic sensor to facilitate the identification of chemicals that can disrupt transgenerational RNA silencing (Galford & Jose, 2020). 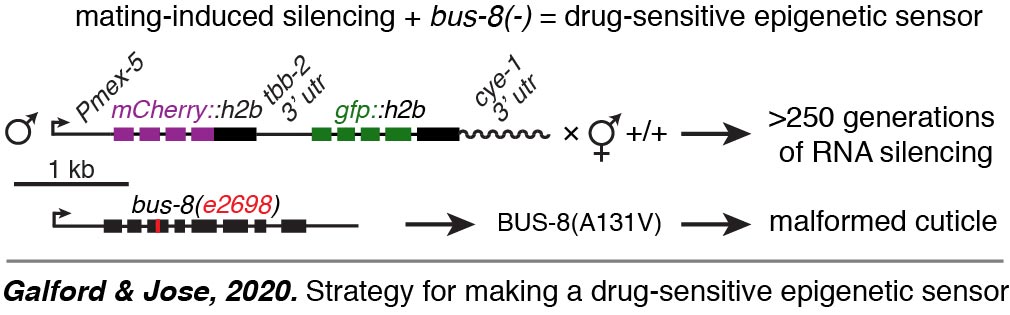 A preliminary screen using 1443 FDA-approved compounds did not reveal any that disrupted transgenerational RNA silencing, but identified three drugs that caused obvious morphological changes. These serendipitously discovered changes provide avenues for exploring the biological impact of these drugs that have been collectively prescribed to millions of people.
A preliminary screen using 1443 FDA-approved compounds did not reveal any that disrupted transgenerational RNA silencing, but identified three drugs that caused obvious morphological changes. These serendipitously discovered changes provide avenues for exploring the biological impact of these drugs that have been collectively prescribed to millions of people.
Chemotaxis. We developed a chemotaxis assay that can measure the response of freely moving C. elegans (Tasnim et al., 2025). 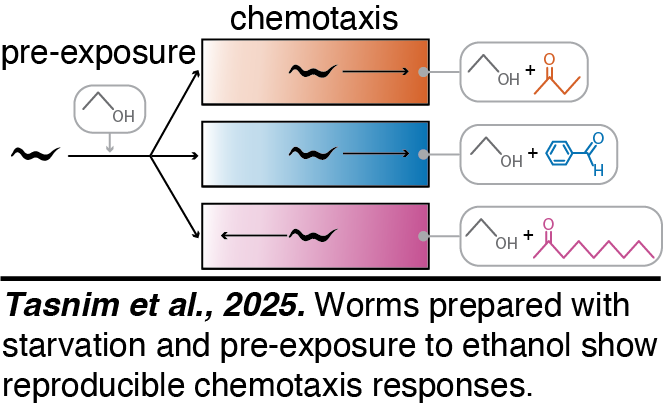 We use opposing orientations of rectangular arenas to control for gradients of unknown cues outside the arena and introduce a measure of dispersal to control for locomotion defects and unknown cues within the arena, enabling the analysis of responses to a variety of chemicals. Using this setup, we found that unfed worms show reproducible movement towards the odorants butanone and benzaldehyde, and away from the odorant nonanone.
We use opposing orientations of rectangular arenas to control for gradients of unknown cues outside the arena and introduce a measure of dispersal to control for locomotion defects and unknown cues within the arena, enabling the analysis of responses to a variety of chemicals. Using this setup, we found that unfed worms show reproducible movement towards the odorants butanone and benzaldehyde, and away from the odorant nonanone.
Back to research
Last updated: Dec 2025