Black - Non-secure connection
Red - Secure connection
Blue - Variations of BLAST showing where the analysis is actually performed
Comparative Bioinformatics (Lab)
Bioinformatics Tools in the Unix Environment
Getting started with unix
Unix is a computer operating system that is widely used for technical computing and development. Although there are proprietary "flavors" of unix (including Apple's Mac OSX, Sun's Solaris, and SGI's Irix), a key feature in the development of this operating system is its generally open source nature. This means that the code that underlies the operating system is available to the users, and thousands of computer scientists working with unix have gradually debugged and expanded unix. Consequently although it is among the oldest operating systems in widespread use, it is also among the most stable, flexible, and powerful of operating systems.
It can also be challenging to the new user. Because unix has been developed by and for computer scientists, it tends to operate with the implicit assumption that you know what you are doing. It is entirely possible to issue a command that will entirely destroy your entire work environment. But it is also possible to use a few commands and accomplish tasks that would take hours of repetitive work on a more "user friendly" operating system. It takes some time and effort to become comfortable with unix, but the rewards are great.
If you have no prior experience with unix, you will have to spend some time becoming familiar with it. There are excellent unix tutorials available on the web. A few of these are:
A university of Maryland tutorial by Charles Lin: http://www.it.umd.edu/unix_tutorial.txt
as well as other tutorials that support CMSC 214: http://www.cs.umd.edu/class/fall2003/cmsc214/Tutorials/
If you don't like that tutorial, try using Google (or some other search engine) to search on "unix tutorial." You will find a large number of alternatives.
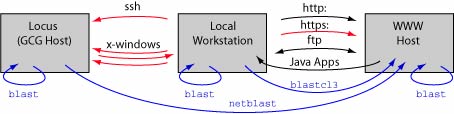
Host/client relationships
We will be using a variety of software tools, some of which are installed on your local workstation, some of which are available on the web, and some of which run on a computer located in UMIACS (the University of Maryland Institute for Advanced Computer Studies), or on other remote computers. To understand what is going on, it will be essential that you understand how these different computers are interacting with each other, and how you are accessing them from your workstation.
 |
Black - Non-secure connection Red - Secure connection Blue - Variations of BLAST showing where the analysis is actually performed |
Note that one in some cases the local workstation is serving primarily to send commands to a remote machine and display the output from those commands. In this case the local workstation may be doing very little work. Some analyses can be quite demanding. How would you decide whether to run such an analysis on a local or a remote computer? Note that it is also sometimes possible for a remote computer to launch a task on your local computer; use caution in such cases as they can present a security risk.
We have already explored some web-based tools at NCBI. Now we will begin to use local tools and tools on a bioinformatics computing platform that is running the software package "GCG."
Open terminal on local machine
Explore the directory structure:
ls
pwd
cd /usr/local/biotools/bin
ls
pwd
cd
pwd
What happened? If you issue the command cd with no arguments it will take you back to your home directory. You can also supply an argument to ls:
ls /usr/local/biotools/bin
What is your current working directory? What is the path to the directory you just listed?
Understanding commands, options, and arguments
Unix will parses one command line at a time; when you press the carriage return key it prompts the computer to interpret what you just typed.
The first string of characters on the line will be interpreted as a command
Many commands have optional settings that can change how the command behaves
Many commands also have arguments. Some arguments are mandatory, others are optional.
Read unix man pages frequently
Use the comand man to look at the arguments for ls and cd. You an exit from a man page by pressing q.
Also look at
%man man