How we think about living systems provides the framework for analyzing experimental results and new experimental discoveries can prompt a revision of such frameworks. By ~2017, we realized that a new framework is required for heredity to explain the inheritance of epigenetic changes. We have since developed this framework progressively from conceptual ideas (cell code) to quantitative explanations (heritable regulatory architectures) of how all regulatory information is transmitted across generations. With these advances, our experimental strategies aim to understand the regulation of two fundamental properties of all living systems - heredity and homeostasis.
Cell Code: Life has diversified since its origin through bottleneck stages that separate successive generations. Each organism develops from the bottleneck stage, typically a single-cell zygote, with two distinct stores of information.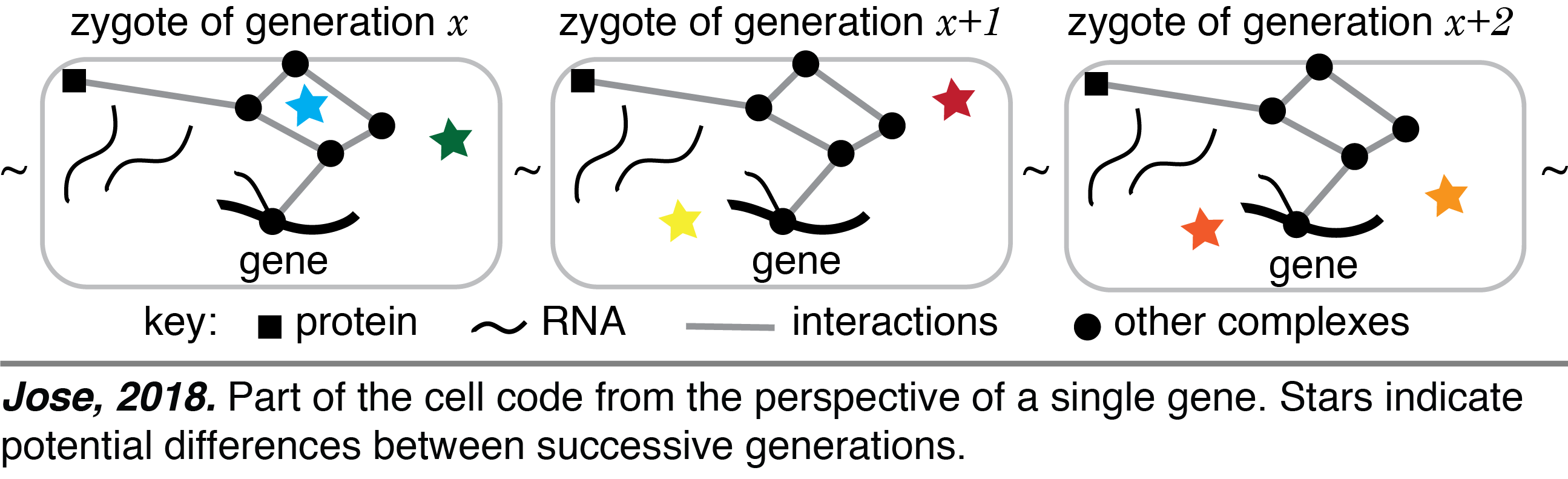 One is the linear DNA sequence that is replicated during cell divisions. The other is the three-dimensional arrangement of regulatory molecules that dictates what is made using the DNA sequence and changes during development but returns to a similar configuration at the start of each generation. These two interdependent stores of information – one replicating with every cell division (e.g., gene sequence) and the other cycling with a period of one generation (e.g., gene regulators) – coevolve and together form the cell code for making an organism (see video). This perspective impacts our understanding of the origins of inherited diseases, the course of evolution, and the synthesis of new life.
One is the linear DNA sequence that is replicated during cell divisions. The other is the three-dimensional arrangement of regulatory molecules that dictates what is made using the DNA sequence and changes during development but returns to a similar configuration at the start of each generation. These two interdependent stores of information – one replicating with every cell division (e.g., gene sequence) and the other cycling with a period of one generation (e.g., gene regulators) – coevolve and together form the cell code for making an organism (see video). This perspective impacts our understanding of the origins of inherited diseases, the course of evolution, and the synthesis of new life.
Entity-Sensor-Property systems: We developed a single framework for parsing all heritable information in living systems. This framework includes cycling stores of information in terms of what an organism can sense about itself and its environment by defining entities, their sensors, and the sensed properties (Jose, 2020a). Entities include small molecules (ATP, ions, metabolites, etc.), macromolecules (individual proteins, RNAs, polysaccharides, etc.), and assemblies of molecules.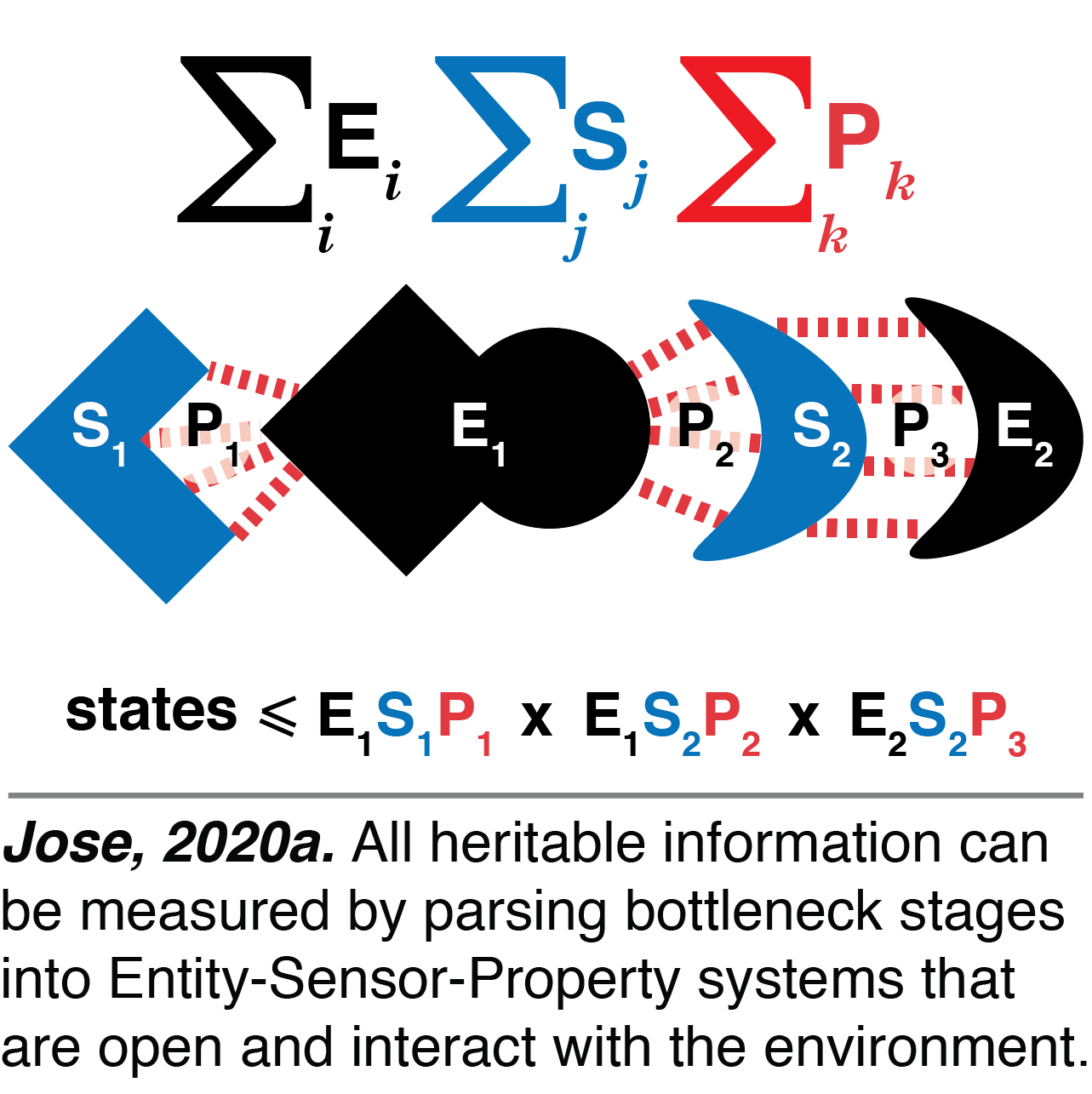 While concentration may be the only relevant property measured by sensors for small molecules, multiple properties that include concentration, sequence, conformation, and modification may all be measured for macromolecules and assemblies. Each configuration of these entities and sensors is thus a potentially vast store of information that drives complex development in each generation. This Entity-Sensor-Property framework explains how sensors limit the number of distinguishable states, how distinct molecular configurations can be functionally equivalent, and how regulation of sensors prevents detection of some perturbations. Overall, this framework is a useful guide for understanding how life evolves and how the storage of information has itself evolved with complexity since before the origin of life.
While concentration may be the only relevant property measured by sensors for small molecules, multiple properties that include concentration, sequence, conformation, and modification may all be measured for macromolecules and assemblies. Each configuration of these entities and sensors is thus a potentially vast store of information that drives complex development in each generation. This Entity-Sensor-Property framework explains how sensors limit the number of distinguishable states, how distinct molecular configurations can be functionally equivalent, and how regulation of sensors prevents detection of some perturbations. Overall, this framework is a useful guide for understanding how life evolves and how the storage of information has itself evolved with complexity since before the origin of life.
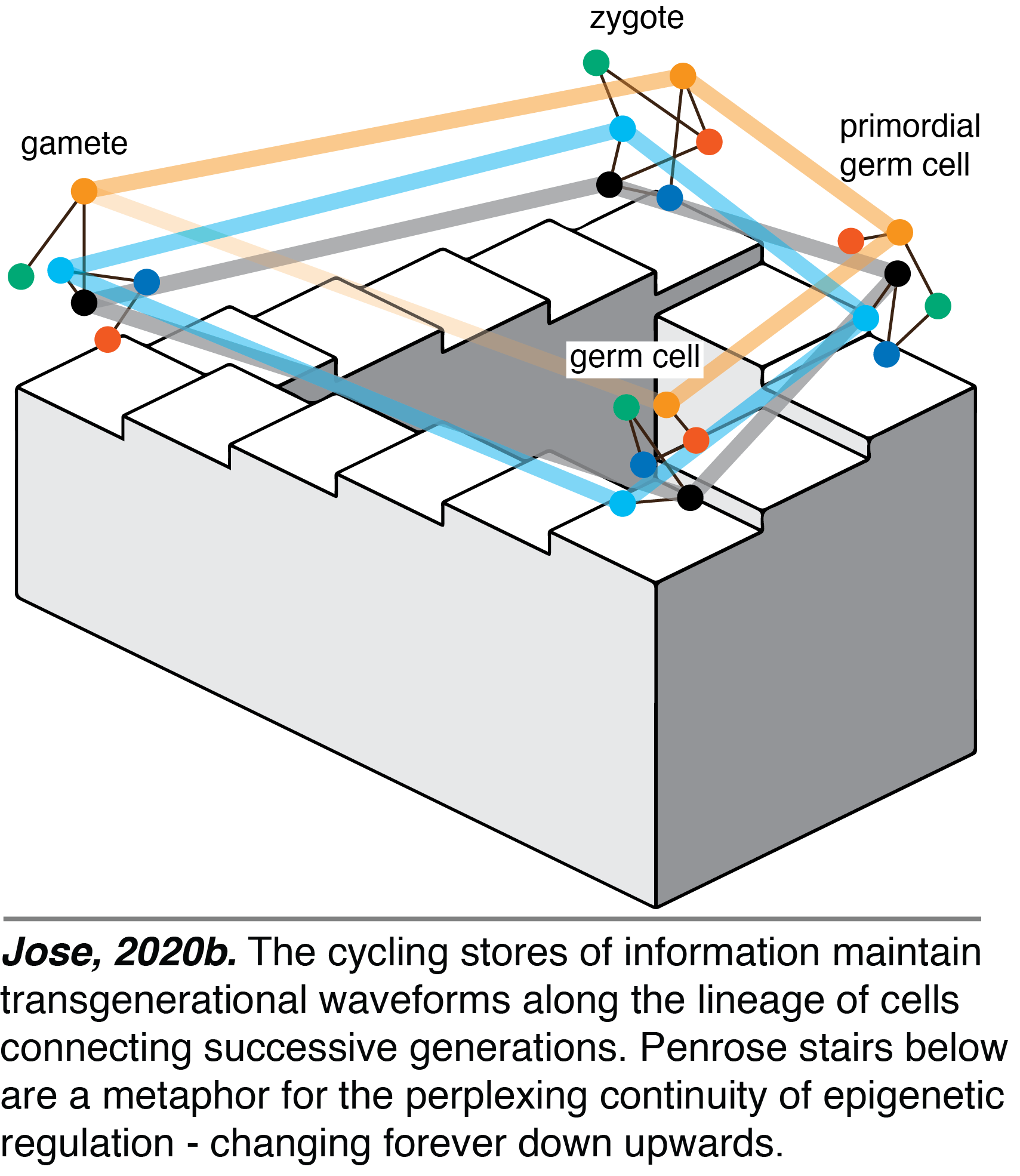
Heritable Epigenetic Changes: We have expanded the conception of heredity to highlight the transgenerational waveforms maintained by cycling stores of information. Unlike heritable genetic changes, which are always associated with mutations in gene sequence, heritable epigenetic changes can be associated with physical or chemical changes in molecules or only changes in the system. The transmission of cycling stores along the continuous lineage of cells that connects successive generations creates waves of activity and localization of the molecules that together form the cell code for development in each generation (Jose, 2020b). As a result, heritable epigenetic changes can include any that can alter a wave such as changes in form, midline, frequency, amplitude, or phase.
Heritable Regulatory Architectures: I. Fundamentals. We have analyzed the simplest heritable regulatory architectures (HRAs) - defining the ABCs of heredity. To persist forever, 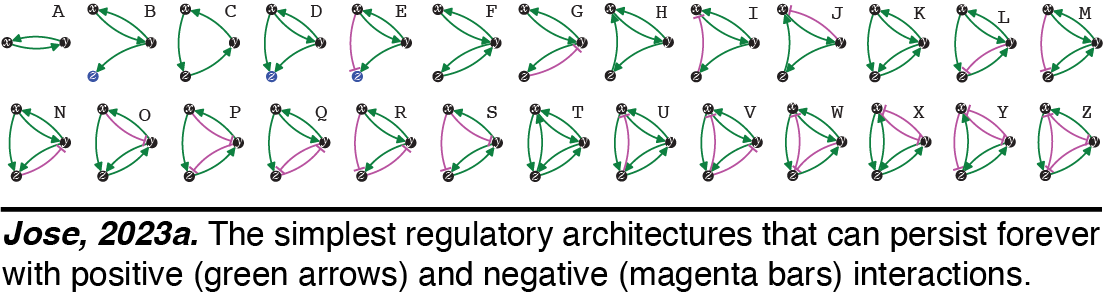 any collection of regulatory interactions needs a perpetual positive feedback loop that promotes the production of every interactor. Using this basic principle and working out the kinds of architectures that are compatible with heredity, we have analyzed the 26 simplest HRAs (Jose, 2023a).
any collection of regulatory interactions needs a perpetual positive feedback loop that promotes the production of every interactor. Using this basic principle and working out the kinds of architectures that are compatible with heredity, we have analyzed the 26 simplest HRAs (Jose, 2023a).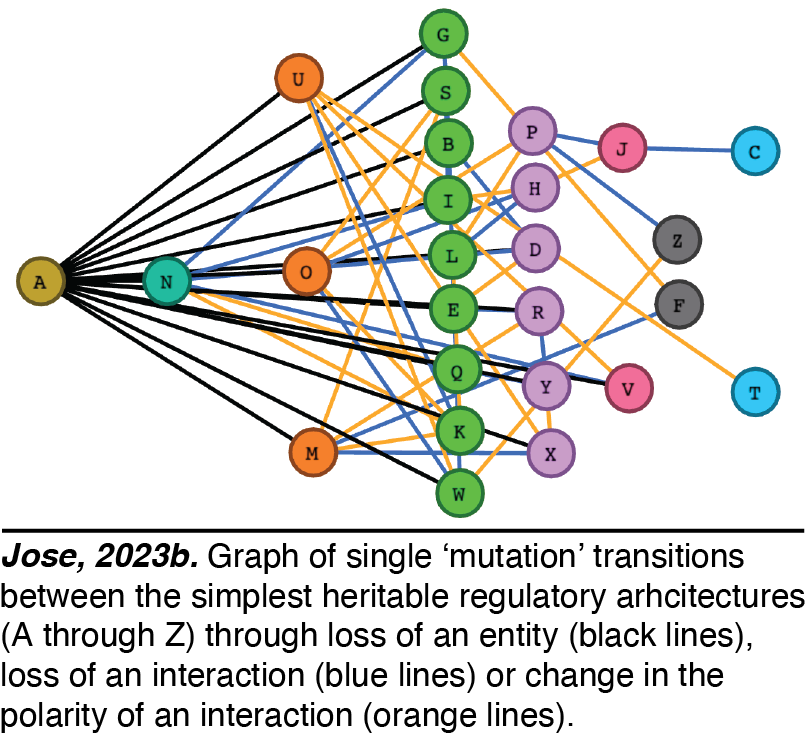 Perturbations of these architectures from steady state provide intuitions for changes that can occur and how they could be useful. For example, continuous growth of a new regulatory architecture after an epigenetic change provides opportunities for achieving new steady states through dilution via cell divisions during development, potentially as part of a new cell type. Transitions from one HRA to another through single changes define possible 'mutations' in the HRA that result in a new state of heritable regulation (Jose, 2023b). We speculate that these constraints could skew the frequencies of different HRAs observed in nature and could restrict the mechanisms available for development and/or evolution.
Perturbations of these architectures from steady state provide intuitions for changes that can occur and how they could be useful. For example, continuous growth of a new regulatory architecture after an epigenetic change provides opportunities for achieving new steady states through dilution via cell divisions during development, potentially as part of a new cell type. Transitions from one HRA to another through single changes define possible 'mutations' in the HRA that result in a new state of heritable regulation (Jose, 2023b). We speculate that these constraints could skew the frequencies of different HRAs observed in nature and could restrict the mechanisms available for development and/or evolution.
Heritable Regulatory Architectures: II. Simulations. Since all collections of regulatory interactors can be abstracted as Entity-Sensor-Property (ESP) systems, we have developed interactive simulations of ESP systems of arbitrary configurations to gain intuitions.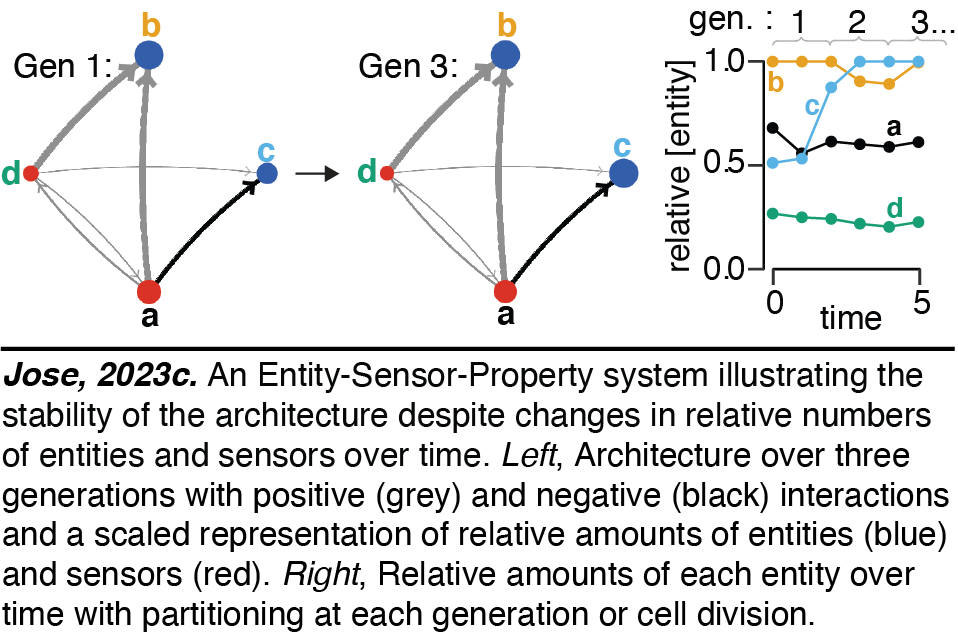 The regulatory interactions can be maintained even as entities and/or sensors change in relative amounts over time (Jose2023c). These simulations enable explorations of the impact of changing many different parameters on the stability of regulatory architectures. These include parameters that control the setup and running of randomly generated ESP systems, the duration of the cell cycle, the maximum number of molecules that will arrest growth until dilution through cell divisions to simulate depletion of raw materials or energy, the maximum number of molecules in total reflecting the limited space occupied by living systems, etc.
The regulatory interactions can be maintained even as entities and/or sensors change in relative amounts over time (Jose2023c). These simulations enable explorations of the impact of changing many different parameters on the stability of regulatory architectures. These include parameters that control the setup and running of randomly generated ESP systems, the duration of the cell cycle, the maximum number of molecules that will arrest growth until dilution through cell divisions to simulate depletion of raw materials or energy, the maximum number of molecules in total reflecting the limited space occupied by living systems, etc.
Heritable Regulatory Architectures: III. Applications. ESP simulations can be applied to understand all regulatory interactions while considering heritability rigorously. 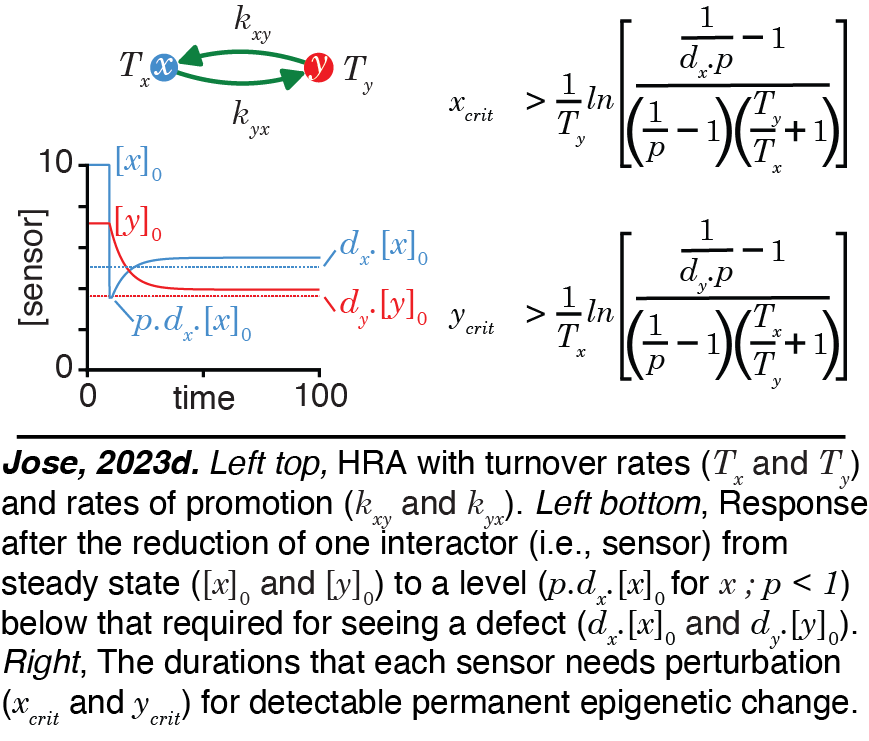 For example,interactive simulations for the regulation of gene expression within the germline in C. elegans can be developed to refine inference as more experimental data are acquired. Requirements for change in the positive feedback loop promoting the persistence of the simplest HRA along with analytical expressions for outcomes after epigenetic perturbations can be deduced (Jose, 2023d). These concepts can be used for quantitative predictions of how changes in measurable RNA intermediates such as 22G RNAs and pUG RNAs generated during heritable RNA silencing in C. elegans impact the durations of such transgenerational epigenetic inheritance.
For example,interactive simulations for the regulation of gene expression within the germline in C. elegans can be developed to refine inference as more experimental data are acquired. Requirements for change in the positive feedback loop promoting the persistence of the simplest HRA along with analytical expressions for outcomes after epigenetic perturbations can be deduced (Jose, 2023d). These concepts can be used for quantitative predictions of how changes in measurable RNA intermediates such as 22G RNAs and pUG RNAs generated during heritable RNA silencing in C. elegans impact the durations of such transgenerational epigenetic inheritance.
Homeostasis: I. Protein-protein interactions from RNA changes. The prevelance of homeostasis suggests that living systems have likely evolved ways to compensate after perturbations to preserve function. If true, this suggests a strategy for identifying protein-protein interactions of regulatory importance, which are difficult to determine experimentally, using RNA changes, which are easily measured (Lalit & Jose, 2025).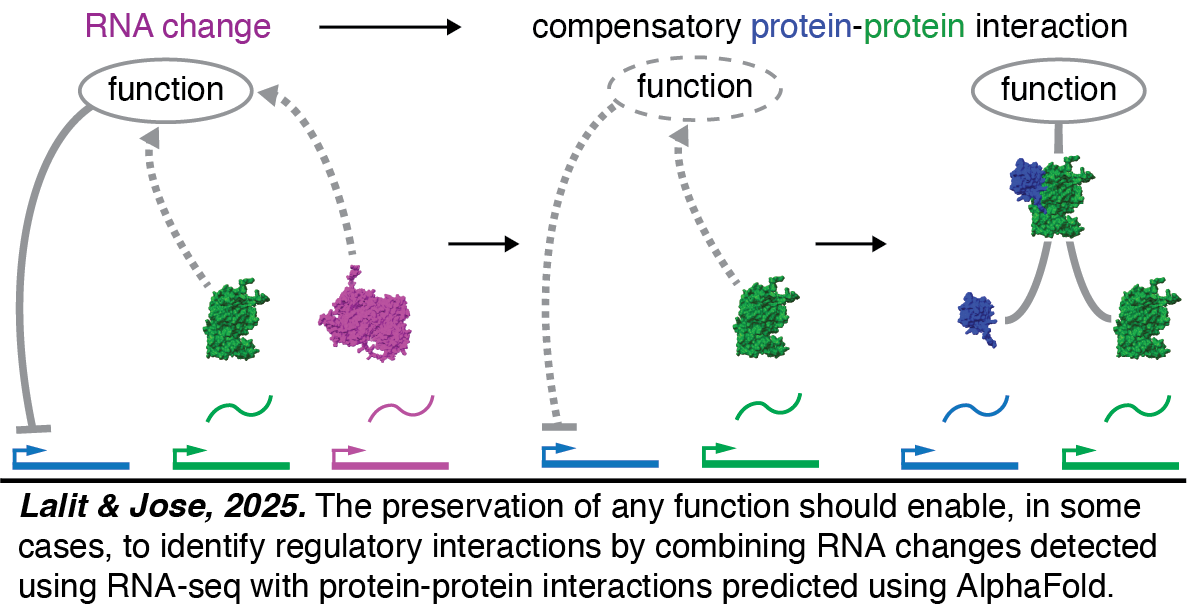 Every field, including RNA silencing in C. elegans, has generated many datasets that document RNA changes after perturbations. The recent availability of AlphaFold and other programs for predicting protein-protein interactions computationally enabled us to identify numerous candidate interactions for testing this hypothesis. Future experimental work will establish whether this strategy is useful.
Every field, including RNA silencing in C. elegans, has generated many datasets that document RNA changes after perturbations. The recent availability of AlphaFold and other programs for predicting protein-protein interactions computationally enabled us to identify numerous candidate interactions for testing this hypothesis. Future experimental work will establish whether this strategy is useful.
Last updated: Dec 2025