- Familiarity with the Unix family of operating systems.
- Familiarity with BLAST.
- Comfort with the world wide web.
Explore the integrated databases provided at NCBI with the "entrez" search tool in order to discover and learn how to navigate the various types of information available.
Three primary consortia provide public access to a tremendous cache of
current scientific knowledge including such disparate topics as sequence
data, protein structure inferences, phylogenetic information, archived
literature, and myriads of accessible tools:
NCBI
EMBL
DDBJ
Each consortium provides purposefully redundant data and separate methods
of accessing the tremendous quantities of information available. For the
purposes of our discussion, we will limit ourselves to examining the tools
available at NCBI.
We have used BLAST to find matches to a query sequence in the GenBank database that is maintained by NCBI. In this exercise we will use Entrez to explore GenBank and look at other resources provided by NCBI. Entrez is an important integrated database search tool provided by NCBI.
NCBI maintains extensive documentation regarding entrez including:
The Help Documentation
and The Entrez Tutorial
Find a gene of interest. If there are very few similar sequences, this is very easy. For example, it would seem that not much of anyone really cares about the (very pretty) green alga Draparnaldia.
Go to the NCBI web site and use Entrez to search GenBank with the name Draparnaldia.
To do this you can just go to NCBI, type draparnaldia in the search box,
be sure that GenBank is selected in the pop-up menu, and click on
go.
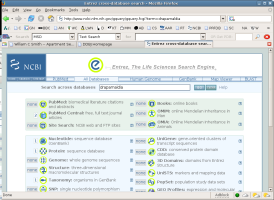
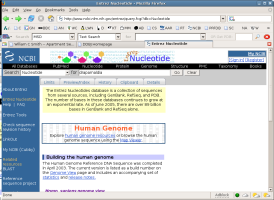
What have you found?
What gene sequences are available for Draparnaldia?
Follow the link and examine the GenBank file. This is a hyperlinked
version of the GenBank flat file format. Explore the links
provided, paying attention to the Taxon browser so that you might
discover other related organisms. Clicking upon the Medline and PubMed
links provides you links to the paper(s) relating to the sequence(s) under
examination. Continue exploring the information provided.
Using the information provided, describe for yourself the characteristics
determined thus far for a transcript from Draparnaldia. What regions of
the Draparnaldia sequence would you expect to be the most conserved?
It is possible to expand the range of sequences found by adding a 'glob' to the end of a search term: compare a search for Draparnaldia to one for "Draparn*", note though that *naldia is not supported.
NCBI has information on the flat file format and other formats used by
GenBank. A great deal of additional information is available on the NCBI
website. Examine some of the other formats provided by clicking upon
the drop down box beside 'Display.'
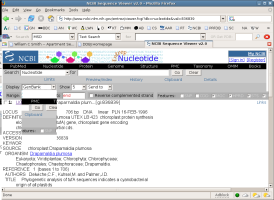
ASN.1 (or for actual information, go
here.), XML, and the Fasta
formats are of special interest. ASN.1 is the format used by NCBI, XML
is a waxing format, and Fasta is a simpler format used by many commonly
used tools.
Follow the link next in the Medline field. What does it show you?
Return to the flat file.
There are two sequences shown at the bottom of the file. How are they
related to each other? Which of these is primary data, and which is
secondary? What does that mean?
Unfortunately the size of the database makes most such searches much more
complex than finding Draparnaldia sequences. Try searching for
genes from the plant Lycopersicon. Notice that you are now
using entrez set to search the nucleotide database; if you want to
explicitly limit your search to the nucleotide database from the outset,
you can select entrez from the menu bar on the NCBI home page,
and then select nucleotide from the entrez menu bar.
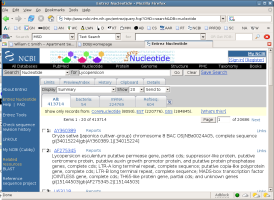
How did your search compare with the search on Draparnaldia?
What is Lycopersicon?
Is there a tufA sequence available for Lycopersicon?
How can you find out?
This is not as easy as it sounds. Try searching on lycopersicon AND tufA
What did you find? Why?
When you are performing an unrestricted search, entrez will find a record if it has the matched words anywhere in the record. Consequently successful searches need to be carefully constructed to find the features that you really want.
You can limit your search to specific fields by entering the search term
in the format
term [field] OPERATOR term [field]
The possible search fields you may search include all the keyed fields of
the ncbi database; listed in the
NCBI help documentation
Entrez permits the use of boolean operators: AND, OR, and NOT are all supported. The boolean operator must be entered in capital letters, but the search terms themselves are not case sensitive. For more information on how entrez handles boolean operators, you can click on entrez nucleotide help, and look for the section on refining your search.
What other elongation factors are available in genbank? Search on
elongation factor.
Is this better or worse? Why?
When you use two or more words in a search, they are combined by default
with AND.
This search will show you several qualitatively different types of sequence. Identify some of the categories of sequence you have found.
You can limit your search to specific types of sequences. Click on the button that says limits, look at what your options are, click on the check box that says all of the above, and then repeat the search.
How has this affected the results of your search?
This is better, but it still isn't giving us the information that we want.
Entrez also maintains a history of your search, and allows you to combine
and modify previous searches. Click on the button that says
history, and examine the information that appears. What you want
to do is combine the search on Lycopersicon with the
limited search on elongation factor. You can do this by referring
to the elements in your history by number. Compose a search, and notice
that there is a new button that says preview. Once your search has
been composed, click on preview and see what happens. If you search is
composed correctly you should have roughly 20 sequences. Once your search
is composed correctly, perform the search and look at the files retrieved.
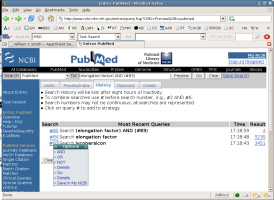
How could you improve this search?
The gene encoding the protein triose-phosphate isomerase (TPI) has been
used in several studies of intron evolution. The distribution of introns
in mosquito TPI genes has been of particular interest. We would like to
identify what TPI genes have been determined from the insect order
Diptera (flies).
How many TPI genes from Diptera can you find?
How many of these are from flies other than Drosophila?
How does the intron content of these genes compare?
We will spend a lot of time examining sequences in the Genbank flat file format; thus we will take a moment to examine it.
LOCUS
-
Includes a unique identifier (often the accession number), and information on the length of the sequence, its data type, and modification date
DEFINITION
-
Provides brief information about the biological attributes of the record. It should include the genus and specific epithet of the source organism, the product name and gene abbreviation, and information about the nature of the sequence.
ACCESSION
-
This is a unique number associated with the record. This number will always be associated with the record, and is the number that you should record to keep track of which records you have worked with.
NID and VERSION
-
These identifiers help keep track of changes that have been made to a record. For a number of reasons, a record may be modified. In this case, the accession number will remain the same, but tracing the history of changes is possible with information from these lines.
KEYWORDS
-
Keywords should be selected from a list of standardized terms. Unfortunately this has not always been the case, and the keywords entry is not always useful.
SOURCE ORGANISM
-
Contains information about what organism the sequence was determined from, as well as taxonomic information about that organism. This is far more complex than it might seem at first, but Genbank is making a concerted effort to have this information be consistent and useful. The Genbank taxonomy browser is a very useful tool available on the Genbank web site.
REFERENCE
-
Information on where the sequence was published. Some records will have several references associated with them. All Genbank entries are required to have at least one reference.
FEATURES
-
The feature table includes a number of sub-headings, and often has extensive information about the biology of the sequence. In the NCBI model, features refer to specific parts of the sequence, while descriptors refer to the entire sequence. The feature table can contain a great deal of information, and if carefully constructed by the original author can be tremendously helpful. Unfortunately they can also contain useless or incorrect information, and some complex sequences are barely annotated.
Gibas and Jambeck, Chapter 6.
Baxevanis & Ouelette, Chapter 5.
http://www.ncbi.nlm.nih.gov:80/entrez/query/static/help/helpdoc.html
Created: Wed Sep 15 00:58:22 EDT 2004 by Charles F. Delwiche
Last modified: Mon Nov 8 15:49:44 EST 2004
by Ashton Trey Belew.